Overview
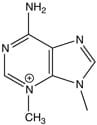
 DNA damage is known to drive mutations and cell toxicity, both of which promote cancer and aging. Recent results also suggest that DNA damage may modulate disease progression following infection. Research in the Engelward laboratory centers on the interplay between DNA damage and its downstream consequences, with the goal of understanding the underlying mechanisms that drive cell toxicity, mutagenesis, and ultimately disease.
DNA damage is known to drive mutations and cell toxicity, both of which promote cancer and aging. Recent results also suggest that DNA damage may modulate disease progression following infection. Research in the Engelward laboratory centers on the interplay between DNA damage and its downstream consequences, with the goal of understanding the underlying mechanisms that drive cell toxicity, mutagenesis, and ultimately disease.
 Progress in science depends on having the appropriate technology to ask the important questions. Recognizing that homology directed repair modulates disease susceptibility, this laboratory was the first to create a ‘recombomouse’ model in which rare recombinant cell fluoresce. This gave rise to many studies of environmental and genetic factors that modulate the risk of large-scale sequence rearrangements.
Progress in science depends on having the appropriate technology to ask the important questions. Recognizing that homology directed repair modulates disease susceptibility, this laboratory was the first to create a ‘recombomouse’ model in which rare recombinant cell fluoresce. This gave rise to many studies of environmental and genetic factors that modulate the risk of large-scale sequence rearrangements.
 With an interest in focusing on common human conditions, we turned our attention to inflammation. By inducing inflammation in the recombomice, we have shown that repeated exposure to inflammation induces genetic rearrangements. Furthermore, recent studies are aimed at understanding the impact of inflammation on susceptibility to other exogenous exposures, such as chemicals in our food or in pollution.
With an interest in focusing on common human conditions, we turned our attention to inflammation. By inducing inflammation in the recombomice, we have shown that repeated exposure to inflammation induces genetic rearrangements. Furthermore, recent studies are aimed at understanding the impact of inflammation on susceptibility to other exogenous exposures, such as chemicals in our food or in pollution.
 The interplay between DNA repair pathways can sometimes be a major driver of sequence instability. We have exploited genetically engineered mice that carry defects in specific DNA repair genes in order to learn about how one DNA pathway impacts another. Interestingly, these studies show that many spontaneous sequence rearrangements are caused by inter-pathway interactions.
The interplay between DNA repair pathways can sometimes be a major driver of sequence instability. We have exploited genetically engineered mice that carry defects in specific DNA repair genes in order to learn about how one DNA pathway impacts another. Interestingly, these studies show that many spontaneous sequence rearrangements are caused by inter-pathway interactions.
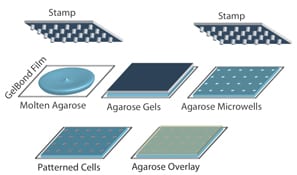
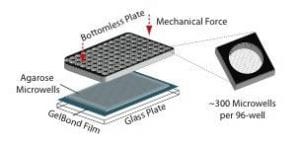
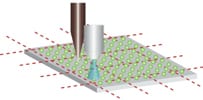 While studies using mouse models have been very informative, we also wanted to develop better ways to study DNA damage and repair in humans. To study DNA damage and repair in human cells, an approach was envisioned for high throughput analysis of DNA damage in human cells. In collaboration with the laboratory of Dr. S, Bhatia (MIT HST EECS), a traditional DNA repair assay called the ‘comet assay’ was modernized by exploiting microfabrication techniques. The resulting DNA damage and repair platform provides better reproducibility and greatly increased throughput. It is our hope that this tool can be used for many applications, including studies of environmental epidemiology.
While studies using mouse models have been very informative, we also wanted to develop better ways to study DNA damage and repair in humans. To study DNA damage and repair in human cells, an approach was envisioned for high throughput analysis of DNA damage in human cells. In collaboration with the laboratory of Dr. S, Bhatia (MIT HST EECS), a traditional DNA repair assay called the ‘comet assay’ was modernized by exploiting microfabrication techniques. The resulting DNA damage and repair platform provides better reproducibility and greatly increased throughput. It is our hope that this tool can be used for many applications, including studies of environmental epidemiology.
 It is increasingly apparent that microbes influence our health in many ways. For example, pathogenic microbes can promote cancer. To learn more about the underlying mechanism of pathogenicity, we are exploring the potential for pathogenic microbes to damage DNA. Interestingly, we have found that S. pneumoniae can induce DNA damage in human cells, and we have contributed to work showing that colibactin, an E. coli pathogenicity factor, is a DNA crosslinking agent. In addition to understanding pathogenicity, our team is also working on understanding the impact of beneficial microbes on cancer risk.
It is increasingly apparent that microbes influence our health in many ways. For example, pathogenic microbes can promote cancer. To learn more about the underlying mechanism of pathogenicity, we are exploring the potential for pathogenic microbes to damage DNA. Interestingly, we have found that S. pneumoniae can induce DNA damage in human cells, and we have contributed to work showing that colibactin, an E. coli pathogenicity factor, is a DNA crosslinking agent. In addition to understanding pathogenicity, our team is also working on understanding the impact of beneficial microbes on cancer risk.
Taken together, work in this laboratory is at the interface between biological engineering and environmental health, with the goals of developing novel technologies, applying these technologies to accelerate basic research, and ultimately using our understanding of disease processes to inform disease prevention.




 For decades, researchers have measured DNA damage using the single cell gel electrophoresis assay, or comet assay. Originally developed by Singh et al in 1988, the basis for the assay is that damaged DNA migrates more readily than undamaged DNA when electrophoresed. Although effectively used in thousands of publications, the assay nevertheless suffers from sample-to-sample variation and it is highly laborious.
For decades, researchers have measured DNA damage using the single cell gel electrophoresis assay, or comet assay. Originally developed by Singh et al in 1988, the basis for the assay is that damaged DNA migrates more readily than undamaged DNA when electrophoresed. Although effectively used in thousands of publications, the assay nevertheless suffers from sample-to-sample variation and it is highly laborious.

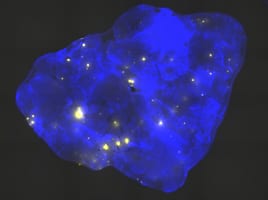 Homologous recombination is critical for repairing double strand breaks (DSBs), but it likely evolved primarily as amechanism for repairing broken replication forks. DNA replication forks can break down when they encounter DNA lesions.
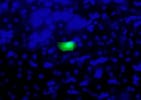
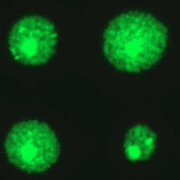
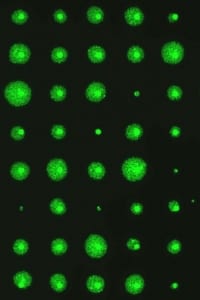
Homologous recombination is critical for repairing double strand breaks (DSBs), but it likely evolved primarily as amechanism for repairing broken replication forks. DNA replication forks can break down when they encounter DNA lesions.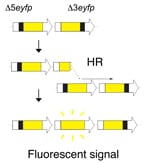 In order to detect homologous recombination in adult animals, we genetically engineered a DNA substrate wherein recombination between two non-functional copies of enhanced yellow fluorescent protein (EYFP) gives rise to fluorescent cells.
In order to detect homologous recombination in adult animals, we genetically engineered a DNA substrate wherein recombination between two non-functional copies of enhanced yellow fluorescent protein (EYFP) gives rise to fluorescent cells.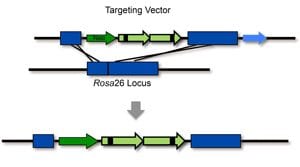
 Recombinant cells can be detected in almost all tissues of the Rosa26 direct repeat (RADR) mice.
Recombinant cells can be detected in almost all tissues of the Rosa26 direct repeat (RADR) mice.